Medical NER and semantic search
Time series forecasting
The github repository :
In this project we were a team of 4 students :
- Mustapha Ajeghrir
- Nouamane Tazi
- Taha Boukhari
- Mohamed Youssef Chouhaidi
The goal of this project was to create a medical NER (named entity recognition), relation extraction between diseases and a semantic search engine.
The NER task :
For this task we have fine tuned several models using HuggingFace like :
- Scibert
- BioBERT
- Electramed
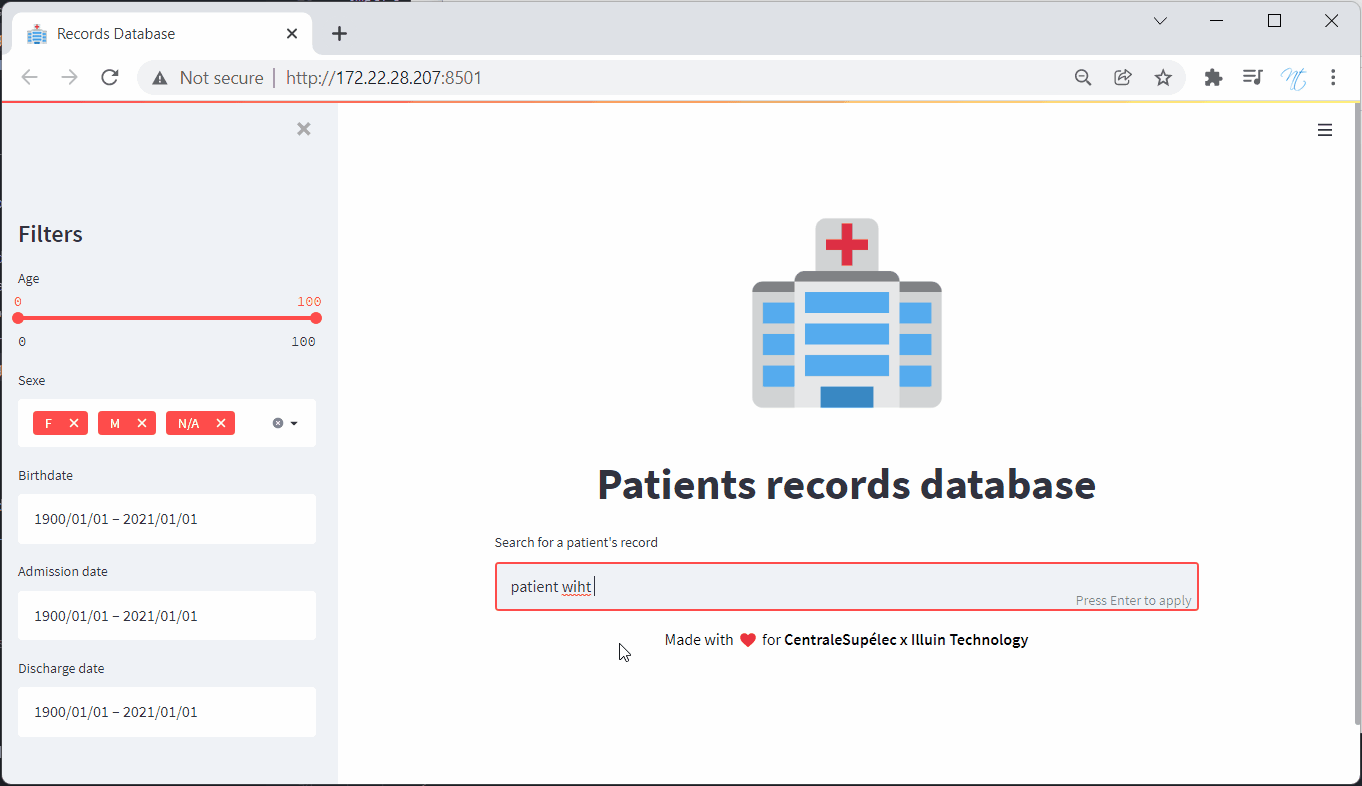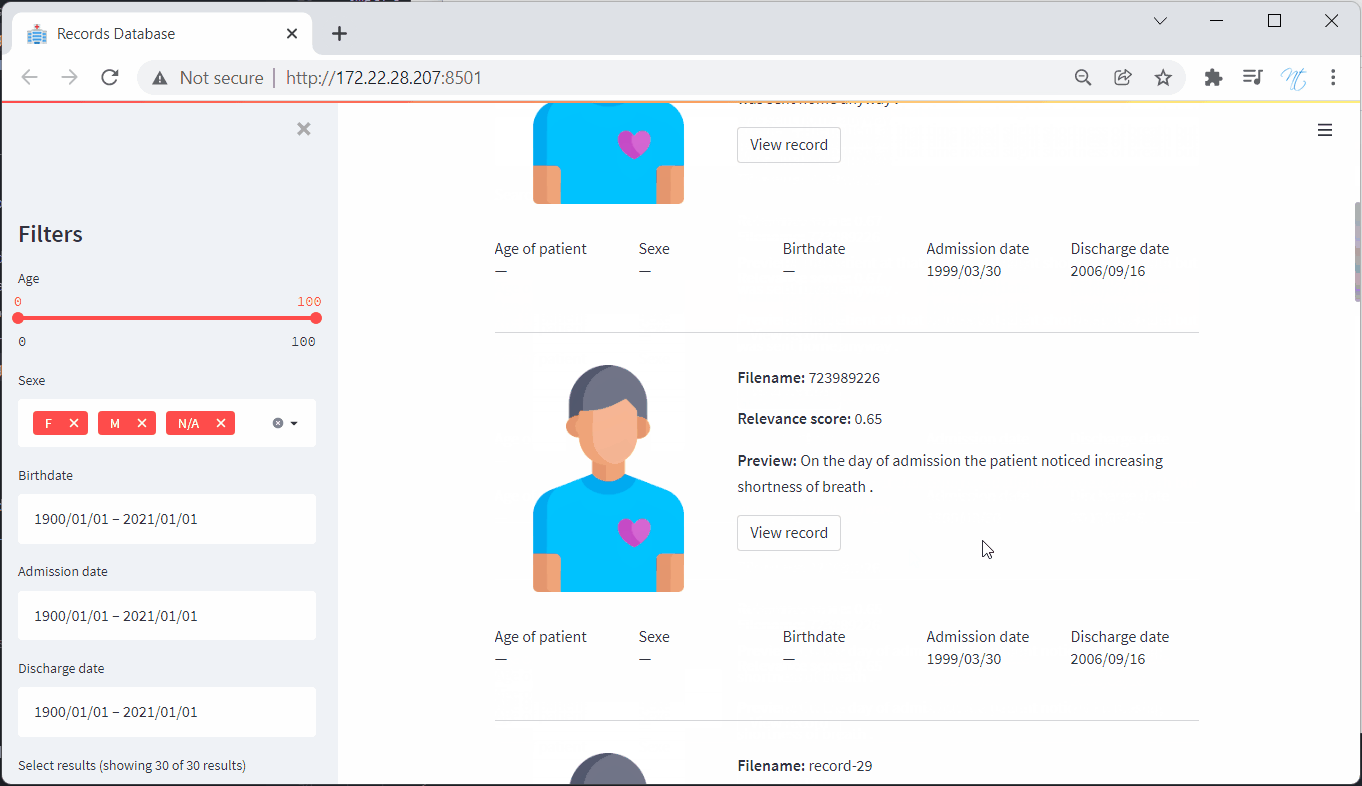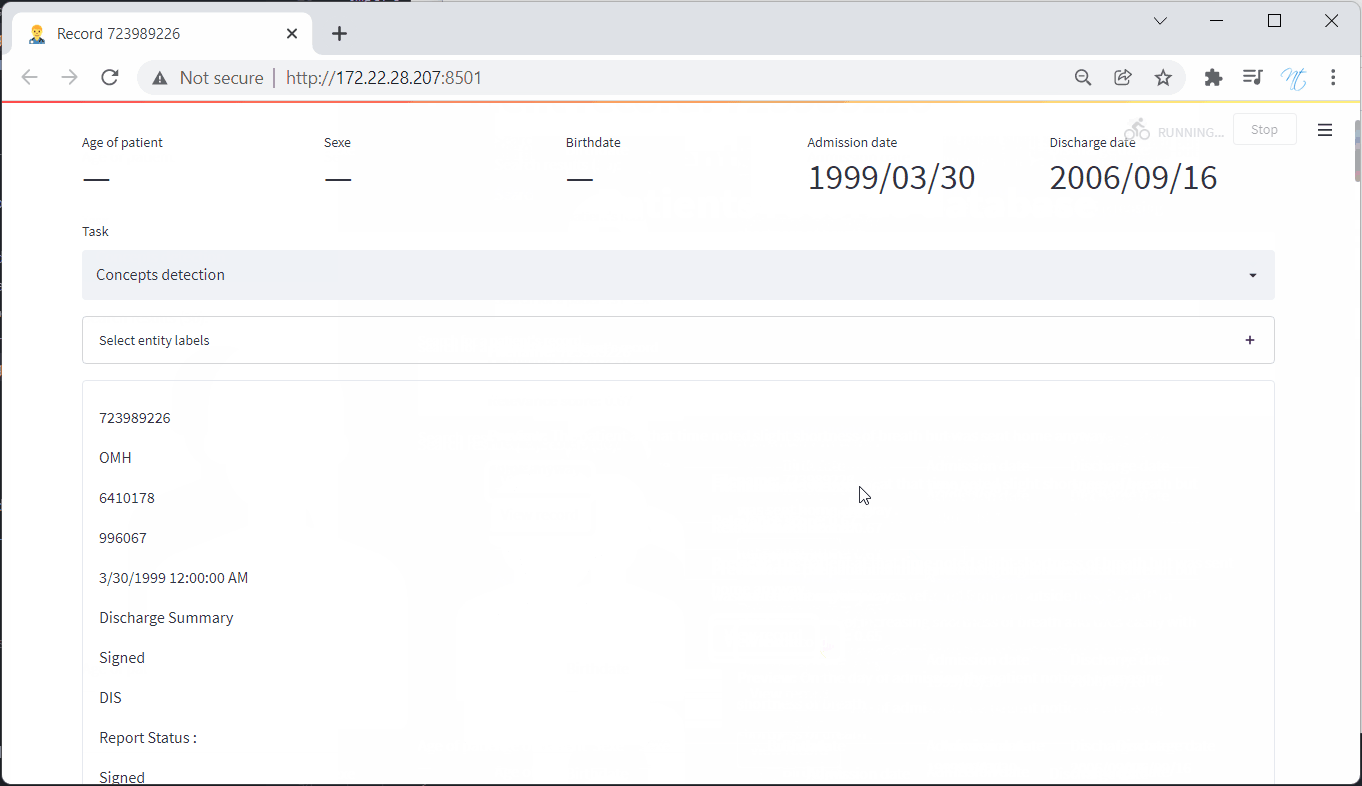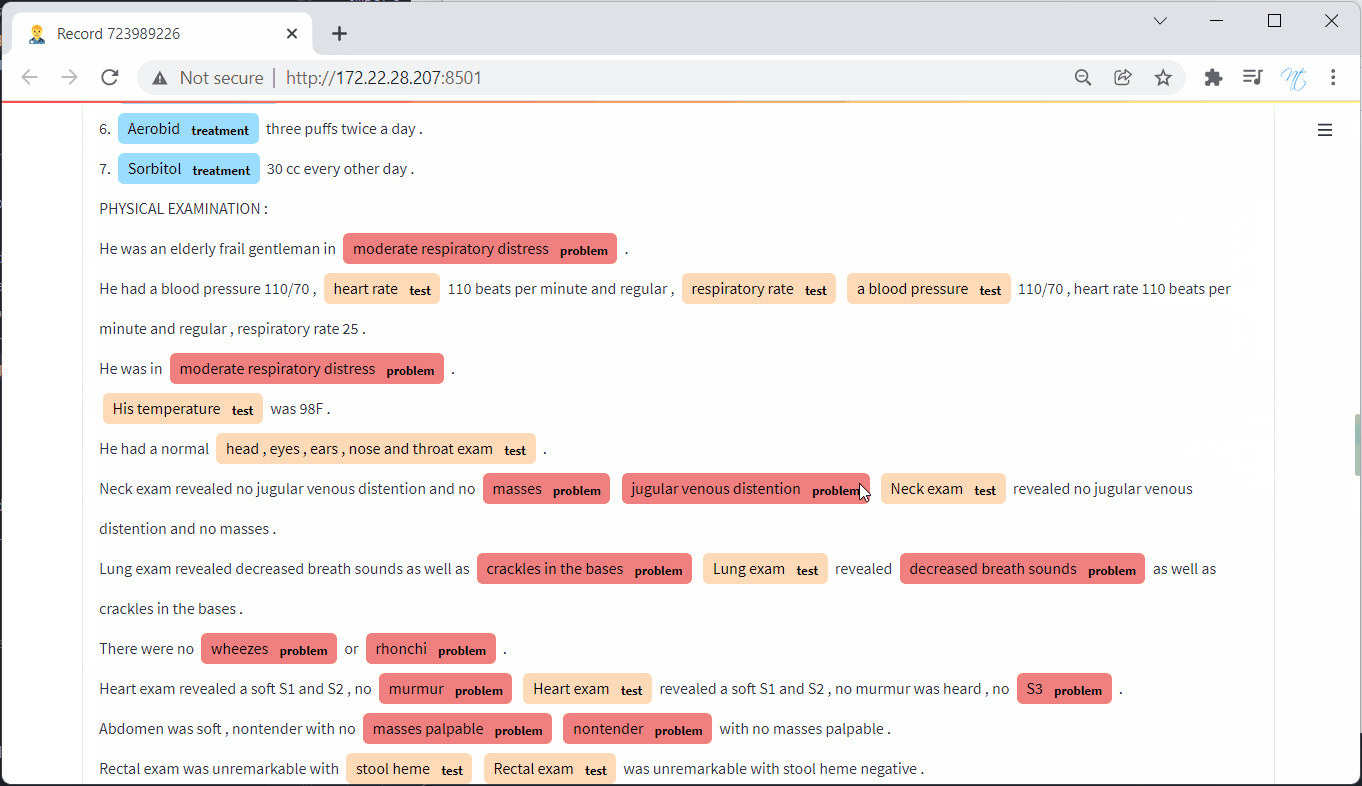
The NER is split into two groups : Concepts and Assertions.
There are 3 Concepts : Test, Treatment, Problem
There are 6 Assertions for problem concepts : Present, Absent, Possible, Conditional, hypothetical, associated
Therefore, we have adopted a Hierarchical Classification :
The Macro average F1-score for the test set was about 0.93 for the concepts and 0.7 for the assertions.
The relation extraction task :
We had to classify the relations between the NER tags, we had 3 groups of relations :
- 1st group : PIP
- 2nd group : TrAP, TrCP, TrNAP, TrIP, TrWP
- 3rd group : TeRP, TeCP
This task relies heavily on the previous one. The NER tags are extracted then for each combition of two concepts, we preprocess the input line by adding << >> for the first concept and [[ ]] for the second concept. For example :
... << C5-6 disc herniation >> with [[ cord compression ]] ...
Then, fine tuned 3 SciBERT models (one for each group of relations) to extract relations between the two concepts.
The semantic search task :
For the semantic search, we have used MiniLM-L6 model to embed the text into a 384 dimension vector space. We then used Annoy to create Indexation trees for fast neighbor search. We finally used Streamlit to visualize the results and Flask as a backend.